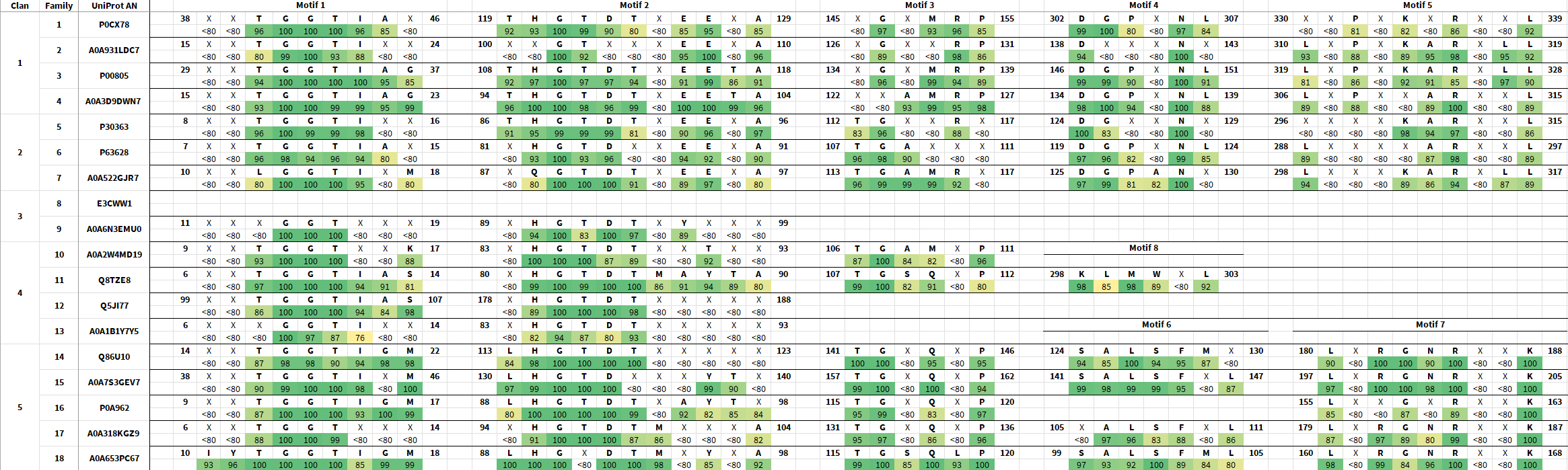
Class 1 l‑asparaginases, previously bacterial-type l‑asparaginases, are the largest and most studied class that contain both l‑asparaginases in pharmaceutical use. Class 1 l‑asparaginases can be found in bacteria, archaea, and eukaryotes, including humans. They display a range of affinities to l‑asparagine, l‑glutamine and sometimes other substrates, such as 1-acyl-sn-glycero-3-phosphocholine or l‑glutamyl-tRNA(Gln), however the l‑asparaginases with the highest affinities to l‑asparagine can be found in this class.
Although sequence similarity can be quite low between sequences, the tertiary structure of the L-asparaginase domain is well conserved throughout the class. This class typically consists of a N-terminal catalytic domain and a C-terminal stabilizing domain connected by a linker, which make up the Class 1 l‑asparaginase monomer. An exception to this are Family 10 l‑asparaginases, which lack the C-terminal stabilizing domain. Family 14 and Family 15 also tend to have additional ankyrin repeats.
Structurally, the l‑asparaginases in this class tend to organize into tetramers and sometimes dimers.