

13 experimentally studied proteins
3 sequences in Swiss-Prot
3,153 unique sequences in UniRef100
Bacterial and archaeal l‑asparaginases
High thermal stability and activity
Dimeric asparagianses from Pyrococcus furiosus and Thermococcus kodakarensis
| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] |
|---|
| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] |
|---|
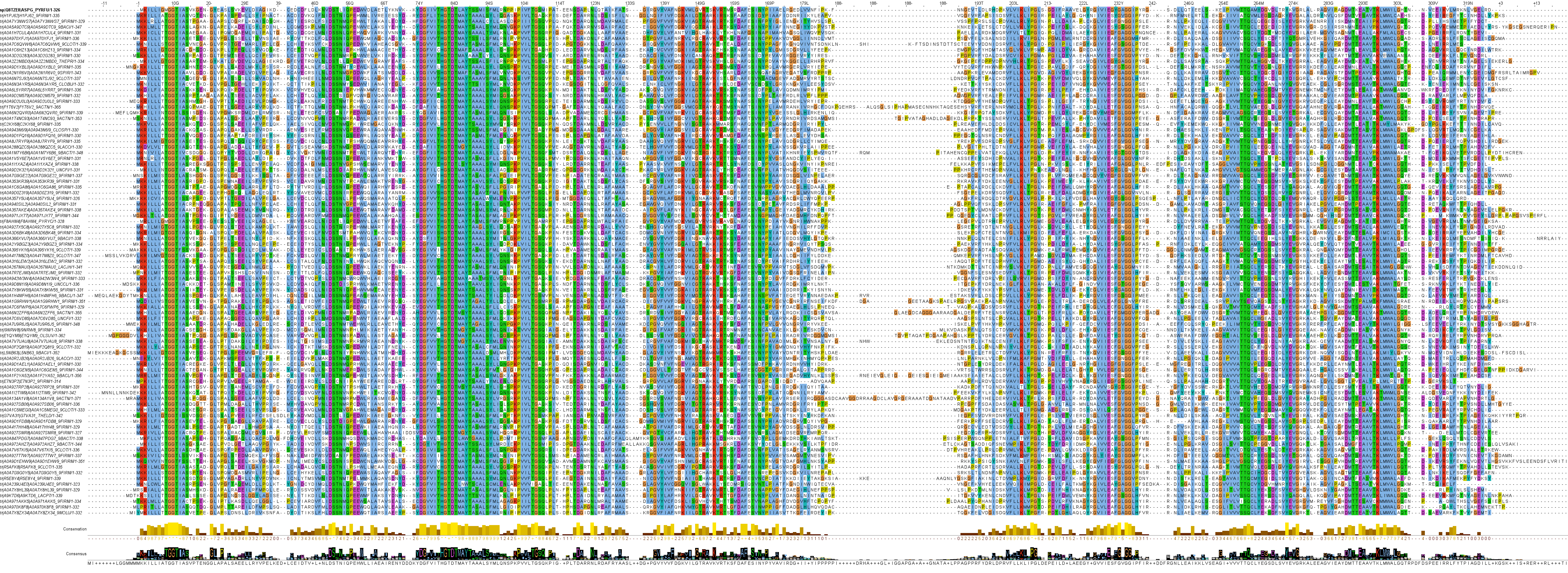
Clan 4 is an interesting clan of bacterial and archaeal l‑asparaginases. Some sequences display characteristics of both "type I" and "type II" l‑asparaginases and some are structurally unique.
Family 11 l‑asparaginases are mostly bacterial, although there are important archaeal representatives present. Three protein structures of l‑asparaginases have been solved in this family, all proteins of archaeal origin: Pyrococcus furiosus PfA (Q8TZE8), Pyrococcus horikoshii PhA (O57797) and the cytoplasmic Thermococcus kodakarensis TkA (Q5JIW4). These l‑asparaginases have a homo dimeric native form and the Km for l‑asparagine tends to be in the millimolar range. These particular l‑asparaginases have likely been chosen for structure analysis due to their archaeal hyperthermophilic origin, but they show high structural similarity (~0.95 TM-score) to numerous bacterial l‑asparaginases in the same family, such as the Bacillus subtilis BsA (P26900).
1Varadi, M et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Research (2024). Licensed under CC BY 4.0.
2Suzek, B.E. et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics (2007). Licensed under CC BY 4.0. Added classification code to sequence headers.
