

24 experimentally studied proteins
12 sequences in Swiss-Prot
11,814 unique sequences in UniRef100
Tetrameric high affinity "type II"
Some enzymes prefer l‑glutamine as a substrate
l‑asparaginases in pharmaceutical use (Escherichia coli, Dickeya chrysanthemi)
| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] |
|---|
| Fam ? Class - Clan - Family | Alt ? Alternative historical name / classification | AN ? UniProt accession number | Name ? UniProt entry name, only given here for Swiss-Prot entries | EC | Organism | Cell-Loc | AAs | Structure | PDB | Km i for Asn [mM] | Vmax i for Asn [μmol/min/mg] | Kcat i for Asn [s-1] |
|---|
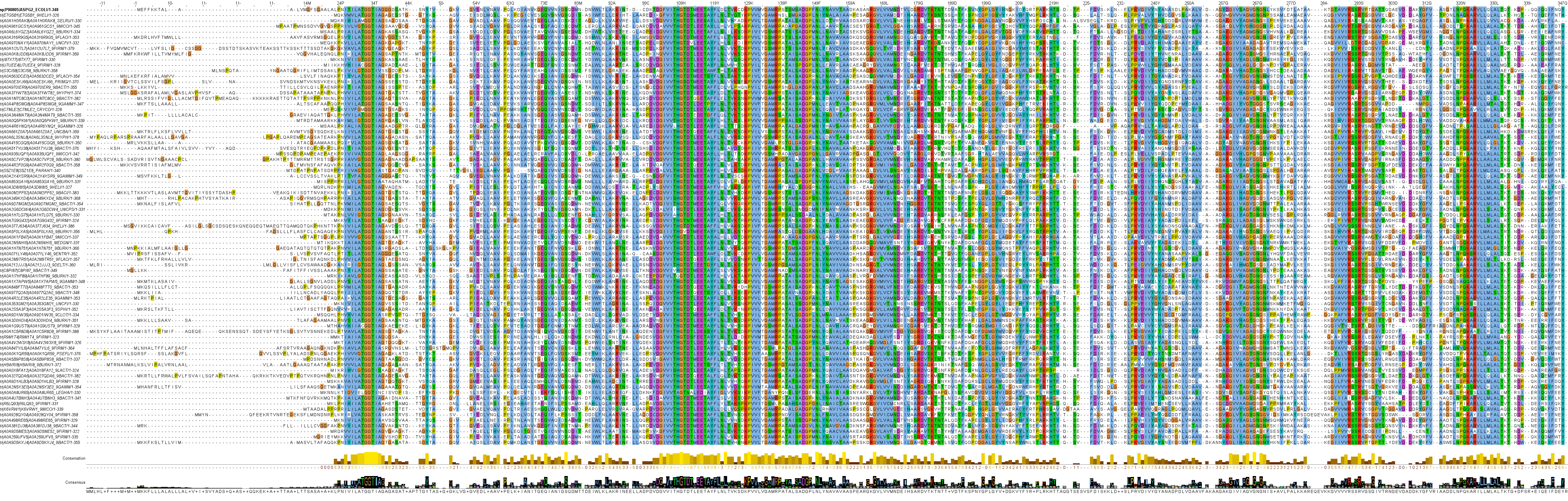
Clan 1 holds 4 families and is somewhat analogous to the historical "type II" l‑asparaginases, as the families of this clan contain "type II" sequences, including the two in therapeutical use. It also has many cytoplasmic sequences and some of these were previously considered to be "type I" l‑asparaginases.
Family 3 has proteins that have been experimentally studied, including the two "type II" l‑asparaginases in clinical use (as the wild-type variants): the Escherichia coli EcAII (P00805) and the Dickeya dadantii ErAII (P06608). Both of these enzymes have a signal sequence, are located in the periplasm, display very high affinity to l‑asparagine and have low, but not negligible, l‑glutaminase activity. This branch also contains many cytoplasmic l‑asparaginases with similarly high affinity to l‑asparagine, such as the Wolinella succinogenes WsAII (P50286) or the allosteric Helicobacter pylori HpAII (Q9ZLB9). However, l‑glutaminase activity can sometimes be higher than l‑asparaginase activity for enzymes in this branch, even though they share high similarity in sequence (~50% identity) and extremely high similarity in structure (~0.97 TM-Score) with EcAII, for example. These enzymes have been named glutaminase-asparaginases (GA), an example would be the Pseudomonas sp. PGA (P10182). Several protein structures have been solved for sequences in Family 3 and these function as homo tetramers.
1Varadi, M et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Research (2024). Licensed under CC BY 4.0.
2Suzek, B.E. et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics (2007). Licensed under CC BY 4.0. Added classification code to sequence headers.
